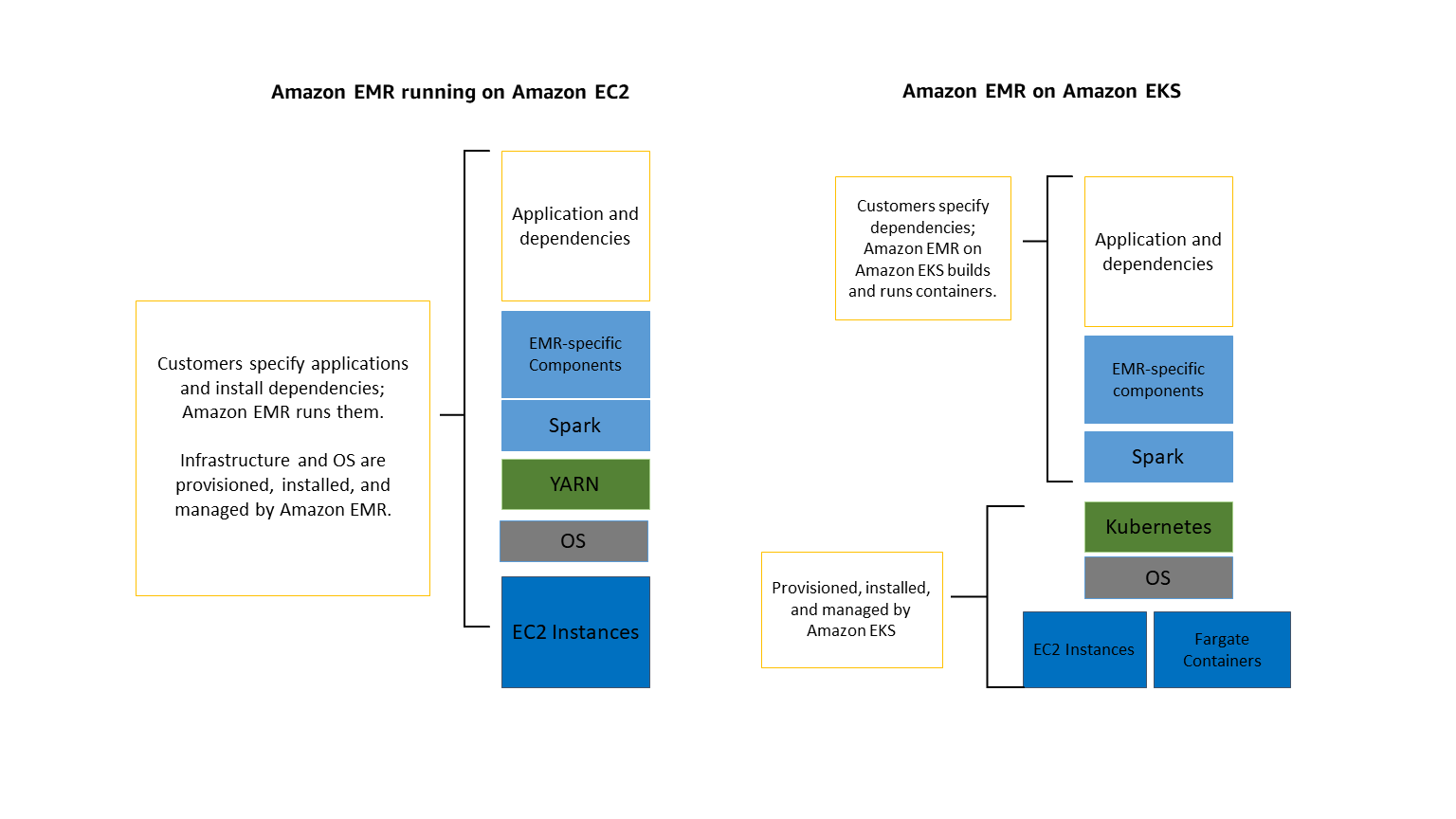
Apache Iceberg is a Lakehouse system with it's key as Metadata.
1. Storage
2. Processing
3. Metadata
Metadata of Metadata
Key Points:
Storage
-
Data files (Parquet, ORC, Avro) are stored in Amazon S3.
-
Iceberg maintains metadata and snapshots to track table versions.
-
ACID Transactions
-
Supports atomic inserts, updates, and deletes directly on S3 data.
-
Prevents read/write conflicts in concurrent jobs.
-
Schema Evolution & Partitioning
-
Allows adding, dropping, or renaming columns without rewriting entire tables.
-
Supports hidden partitioning for efficient queries.
-
Query Engine Compatibility
-
Works with Spark, Flink, Trino, Presto, Athena, and Glue.
-
Enables time travel to query historical snapshots of data.
-
Lakehouse Advantage
-
Combines data lake storage (S3) with data warehouse-like capabilities.
-
Efficient for batch analytics, streaming, and ML pipelines.
🔹 Example Workflow on AWS:
-
Store raw/processed Parquet files in S3.
-
Create an Iceberg table referencing the S3 location.
-
Query and update data using Spark SQL or Athena with ACID guarantees.
-
Enable time travel for auditing or rollback.
Create Iceberg Table in Athena:
CREATE TABLE iceberg_users (
id INT,
name STRING,
event_date DATE
)
PARTITIONED BY = (day(event_date))
LOCATION 's3://your-s3-bucket/iceberg-warehouse/iceberg_users/'
TBLPROPERTIES (
'table_type' = 'ICEBERG',
'write.format.default' = 'parquet'
);
Reference: https://www.youtube.com/watch?v=iGvj1gjbwl0
🔹 Operations using PySpark:
Writing Data to an Iceberg Table
# Initialize SparkSession with Iceberg support
spark = SparkSession.builder \
.appName("IcebergReadWrite") \
.config("spark.sql.catalog.glue_catalog", "org.apache.iceberg.spark.SparkCatalog") \
.config("spark.sql.catalog.glue_catalog.warehouse", "s3://my-iceberg-bucket/warehouse/") \
.config("spark.sql.catalog.glue_catalog.catalog-impl", "org.apache.iceberg.aws.glue.GlueCatalog") \
.config("spark.sql.catalog.glue_catalog.io-impl", "org.apache.iceberg.aws.s3.S3FileIO") \
.getOrCreate()
# Sample DataFrame
data = [
(1, "Alice", 2025),
(2, "Bob", 2024),
(3, "Charlie", 2025)
]
columns = ["id", "name", "year"]
df = spark.createDataFrame(data, columns)
# Write to Iceberg table in Glue catalog (S3 backend)
df.writeTo("glue_catalog.db.iceberg_users").createOrReplace()
Reading from an Iceberg Table