Broadcast variables allow the programmer to keep a read-only variable cached on each machine rather than shipping a copy of it with tasks. They can be used, for example, to give every node a copy of a large input dataset in an efficient manner. Spark also attempts to distribute broadcast variables using efficient broadcast algorithms to reduce communication cost.
Spark actions are executed through a set of stages, separated by distributed “shuffle” operations. Spark automatically broadcasts the common data needed by tasks within each stage. The data broadcasted this way is cached in serialized form and deserialized before running each task. This means that explicitly creating broadcast variables is only useful when tasks across multiple stages need the same data or when caching the data in deserialized form is important.
Accumulators are variables that are only “added” to through an associative and commutative operation and can therefore be efficiently supported in parallel. They can be used to implement counters (as in MapReduce) or sums. Spark natively supports accumulators of numeric types, and programmers can add support for new types.
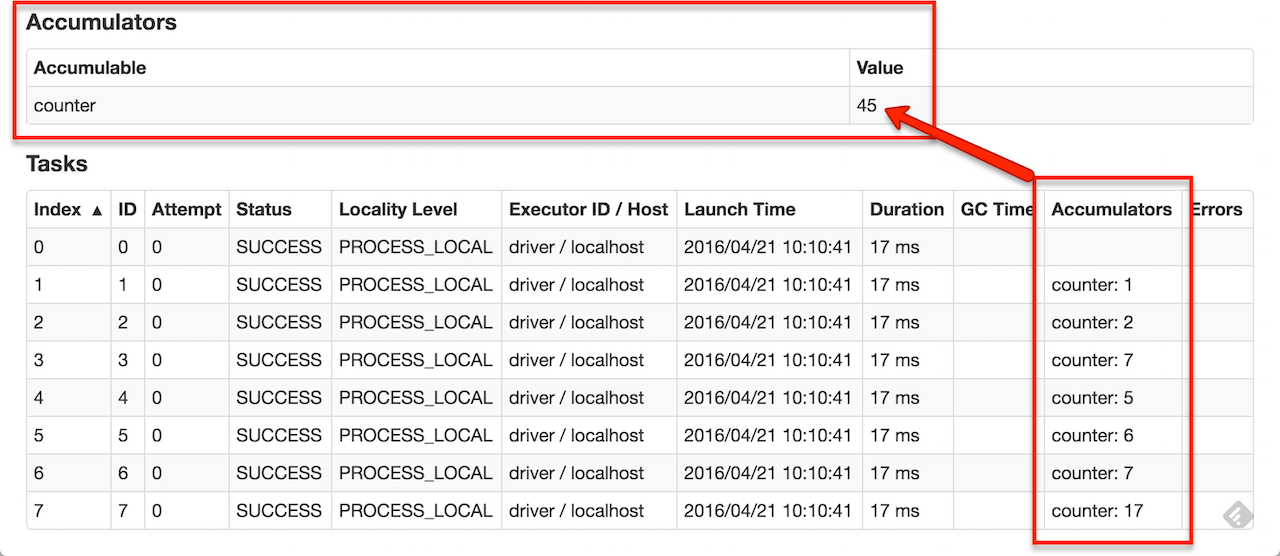
As a user, you can create named or unnamed accumulators. As seen in the image below, a named accumulator (in this instance counter) will display in the web UI for the stage that modifies that accumulator. Spark displays the value for each accumulator modified by a task in the “Tasks” table.
Tracking accumulators in the UI can be useful for understanding the progress of running stages (NOTE: this is not yet supported in Python).
The code below shows an accumulator being used to add up the elements of an array:
scala> val accum = sc.longAccumulator("My Accumulator")
accum: org.apache.spark.util.LongAccumulator = LongAccumulator(id: 0, name: Some(My Accumulator), value: 0)
scala> sc.parallelize(Array(1, 2, 3, 4)).foreach(x => accum.add(x))
...
10/09/29 18:41:08 INFO SparkContext: Tasks finished in 0.317106 s
scala> accum.value
res2: Long = 10
Another real time example where I have used accumulators:
Case: I have written an API that does an upsert to RDBMS which uses DriverManager class where the records are parsed and inserted like insert into ...
During this process if an exception has occured due to improper data in one of the record then we need to capture this exception and mark the function output to represent its failure to upsert. Increment the counter value in the exception block as,
retVal += 1
exceptionReturnedFrmExecutor +=sqlExp.getMessage()
return it in the
finally{
(retVal, exceptionReturnedFrmExecutor)
}
Here, both retVal and exceptionReturnedFrmExecutor are accumulators.
Spark SQL vs Spark Session
Prior to Spark 2.0 there is a need to create a SparkConf and SparkContext to interact with Spark, and then SQLContext.
Whereas in Spark 2.0 the same effects can be achieved through SparkSession, without expliciting creating SparkConf, SparkContext or SQLContext, as they’re encapsulated within the SparkSession. Using a builder design pattern, it instantiates a SparkSession object if one does not already exist, along with its associated underlying contexts.
Eg: val sparkSession = SparkSession.builder.
master("local")
.appName("spark session example")
.getOrCreate()
Difference between spark.jars and spark.driver.extraClassPath
Use --jars if you want to make these jars available to both driver and executor class-paths. If the required jar is only to be used by driver code, use option --driver-class-path
Launching spark-shell with external jars.
spark-shell --jars /usr/lib/hive/lib/json-udf-1.3.7-jar-with-dependencies.jar,/usr/lib/hive/lib/json-serde-1.3.7.3.jar
In Post, http://leelaprasadhadoop.blogspot.in/2017/07/hive-functions.html
a) A JAR has been added to hive
b) Created function to use as UDF.
hive> add jar hdfs:////user/HiveHbase/hive-contrib-1.1.0.jar;
hive> CREATE TEMPORARY FUNCTION row_sequence as 'org.apache.hadoop.hive.contrib.udf.UDFRowSequence';
Same can be acheived in Spark via below commands.
Launch Spark-shell along with the external JAR
spark-shell --jars /home/gorrepat/hive-contrib-1.1.0.jar
scala> import org.apache.spark.sql.hive.HiveContext
scala> val hq = new HiveContext(sc);
scala> hq.sql("""CREATE TEMPORARY FUNCTION row_sequence as 'org.apache.hadoop.hive.contrib.udf.UDFRowSequence'""")
scala> hq.setConf("set hive.mapred.mode","nonstrict") //same as hive> set hive.mapred.mode=nonstrict;
To SEE THE LOADED Classpath JARS,
In Scala ,
val cl = ClassLoader.getSystemClassLoader
cl.asInstanceOf[java.net.URLClassLoader].getURLs.foreach(println)
Inserting data in Hive table from Spark SQL
val data = hq.sql("select \"leela\" as name)
data.write.mode("append").saveAsTable("emp")
OR
hq.sql("insert into table emp select name from student_hive")
Writing data to HDFS without using saveAsTextFile. This helps in case where this API can't be used.
import org.apache.hadoop.conf.Configuration
import org.apache.hadoop.fs.{FileSystem, Path}
import org.apache.hadoop.fs.{FSDataOutputStream, Path}
import org.apache.spark.input.PortableDataStream
import java.io._
def writeAsString(hdfsPath: String, content: String) {
val fs = {
val conf = new Configuration()
FileSystem.get(conf)
}
val path: Path = new Path(hdfsPath)
if (fs.exists(path)) {
fs.delete(path, true)
}
val dataOutputStream: FSDataOutputStream = fs.create(path)
val bw: BufferedWriter = new BufferedWriter(new OutputStreamWriter(dataOutputStream, "UTF-8"))
bw.write(content)
bw.close
}
rdd conversion to DataFrame
There are few options available to convert rdd to DF.
1. Create a case class and map rdd values to case class fields and use toDF().
Note: Spark 1.6 and below had a limitation of not accepting more than 23 fields in a case class.
2. StructType & StructField is a goo way, however need to have the data in the form of Rows and not in Array.
Spark Client mode and Cluster mode
Client mode: The driver resides in the Client machine from which the application is launched vis spark-submit.
Cluster mode: Spark Driver resides in Application Master. Even though we shut down the machine from which spark-submit is triggered, still the application runs because the driver is in Application Master.
Source:
https://www.youtube.com/watch?v=vJ0eUZxF80s
Executors,Stage, partitions and Tasks.
Take a case where Driver has submitted the Job for 2 Executors to execute. Data is in 5 partitions.
In case of 5 partitions a maximum of 5 tasks can be executed, but in this case there are only 2 Executors, so in-parallel only 2 tasks will get execute and the other 3 will wail for one of them to complete and get processed.
Stage: It can be called as a set of transformations execution. When ever a wide transformation is encountered a new stage would be created(eg. Reducebykey or join) in which data shuffles across the partitions. Stages will comprised of Tasks.
Sources:
https://www.youtube.com/watch?v=qctfDbrvQ8s
https://www.youtube.com/watch?v=fyTiJLKEzME
Parallelism in Spark using Futures
follow:
http://www.russellspitzer.com/2017/02/27/Concurrency-In-Spark/#concurrency-in-spark
Sample code for simple reading 2 DF's, Join and Writing the result to a New DB
object stgIngestion {
def main(args: Array[String]): Unit = {
println("Say hello")
val spark = SparkSession.builder
.appName("stgingIngestion")
.enableHiveSupport()
.config("hive.exec.dynamic.partition","true")
.config("hive.exec.dynamic.partition.mode","nonstrict")
.config("spark.sql.parquet.writeLegacyFormat","true")
.getOrCreate()
spark.sparkContext.hadoopConfiguration.set("mapreduce.fileoutputcommitter.algorithm.version", "2")
spark.sparkContext.hadoopConfiguration.set("speculation", "false")
spark.conf.set("spark.sql.autoBroadcastJoinThreshold", -1)
val filename = args(0)
val srcPathTblsPath = args(1)
val targetHiveDB = args(2)
val targethivePath = args(3)
val tableList = Source.fromFile(filename).getLines.toList
val tableIterator = tableList.toIterator.map(table => Future{
val df = spark.read.format("orc").load(s"${srcPathTblsPath}/${table}/")
val df_id = spark.read.format("orc").load(s"${srcPathTblsPath}/${table}_id/")
df.withColumn("row_num", row_number() over(partitionBy(col("id")).orderBy(desc("updatetime")))).
filter(col("row_num") === "1").
join((df_id),Seq("id"),"inner").
write.format("parquet").
options(Map("path" -> s"${targethivePath}/${table}")).
mode(SaveMode.Overwrite).partitionBy("ingestiondt").
saveAsTable(targetHiveDB + "." + table)
})
val timeout: Duration =Inf
// Await.result(Future.sequence(tableIterator),timeout) //Without Sliding window
def awaitSliding[T](it: Iterator[Future[T]], batchSize: Int = 2, timeout: Duration = Inf): Iterator[T] = {
val slidingIterator = it.sliding(batchSize - 1).withPartial(true)
val (initIterator, tailIterator) = slidingIterator.span(_ => slidingIterator.hasNext)
initIterator.map(futureBatch => Await.result(futureBatch.head, timeout)) ++
tailIterator.flatMap(lastBatch => Await.result(Future.sequence(lastBatch), timeout))
}
awaitSliding(tableIterator).foreach(x => println("Done"))
}
}
Sample spark-submit command,
spark-submit --class com.practice.stgIngestion --master yarn --deploy-mode client --conf spark.shuffle.spill=true --conf spark.executor.extraJavaOptions=-XX:MaxPermSize=1024m --conf spark.sql.planner.externalSort=true --conf spark.shuffle.manager=sort --conf spark.ui.port=8088 --conf spark.executor.memoryOverhead=12096 --conf spark.driver.memoryOverhead=12096 --conf spark.rpc.message.maxSize=1024 --conf spark.file.transferTo=false --conf spark.driver.maxResultSize=10g --conf spark.rdd.compress=true --conf spark.executor.cores=5 --conf spark.executor.memory=10g --conf spark.driver.memory=20g --conf spark.executor.instances=2 --jars /home/hadoop/data/sqljdbc42.jar /home/hadoop/data/demo_2.11-0.1.jar
More the Number of executors on Big tables might lead to JAVA Heap space error. So, specified spark.executor.instances=2
Note: The TPS for this Job is about 300k per second. Pulled about 1 billion records from all the 10 parallel threads.
pseudo code for executing functionality with future execution context, handling exceptions, logging status in Hive
function call from main(),
val viewIterator = readWriteToHDFS(spark)
awaitSliding(viewIterator, threadsCount).foreach(x => println("Done"))
Functions Definition,
import scala.concurrent.{ Await, Future }
import scala.concurrent.ExecutionContext.Implicits.global
import scala.concurrent.duration._
def readWriteToHDFS(spark: SparkSession): Iterator[Future[Unit]] = {
spark.read ...
val viewlist = spark.read.format("csv").option("header", true).option("delimiter", ",").load(view_path).collect()
viewIterator = viewlist.toIterator.map { a =>
Future {
try {
....
....
writeAuditEntry(spark, viewName, 0, audithive_tableName, s"Success", "Successfully inserted the records")
}
catch {
case ex: FileNotFoundException => {
println("File not Found")
}
case e: Exception => {
println("Unknown exception occured for ${viewname} : $e")
val stackTraceStr = e.getStackTraceString
logger.error(s"Exception occured for ${viewname} and stackTraceStr is:" + stackTraceStr)
writeAuditEntry(spark, viewName, 0, audithive_tableName, s"Failed", e.getMessage)
}
}
}
}
(viewIterator)
}
def writeAuditEntry(spark: SparkSession, tableName: String, recordCount: Long, hiveTable: String, auditStatus: String, auditMessage: String): Unit = {
val newauditMsg = removeEscapeCharacters(auditMessage)
val insertCmd = s"insert into ${audithive_tableName} values('${tableName}', ${recordCount},'${auditStatus}','${newauditMsg}')"
println("insertCmd = " + insertCmd)
spark.sql(insertCmd)
}
Idiom of dataframe PartitionBy and Coalesce
If there is a usecase to write the dataframe to a location Partitioned by few columns and in each column we would like to have a single parquet file.
This is possible with below line of code, however this takes lot of time and on large set of data the job fails as well.
df.coalesce(1).write.partitionBy("entity", "year", "month", "day", "status").mode.(SaveMode.Append).parquet(s"${location}")
partitionBy - will create partitions and save data accordingly.
coalesce - problem with using coalesce(1) is that your parallelism drops to 1, and it can be slow at best and error out at worst. Increasing that number doesn't help either -- if you do coalesce(10) you get more parallelism, but end up with 10 files per partition.
To get one file per partition without using coalesce(), use repartition() with the same columns you want the output to be partitioned by. So in your case, do this
df.repartition("entity", "year", "month", "day", "status").write.partitionBy("entity", "year", "month", "day", "status").mode(SaveMode.Append).parquet(s"$location")
Coalesce() vs Repartition()
The repartition algorithm does a full shuffle of the data and creates equal sized partitions of data. coalesce combines existing partitions to avoid a full shuffle.
Coalsece:
Avoids full shuffle.
Results in unequal partitions
Repartition:
Full shuffle and altogether creates new partitions.
Almost equal sized partitions.
colorDf = peopleDf.repartition(5)
OR
colorDf = peopleDf.repartition($"color") //This is called Repartition by column and this will ensure that
//all the same columns values are present in the same partition.
Eg: Blue, 1
Blue, 2
Red, 1
Blue,1
This creates 2 Partitions where all the Blue colors will be in a partition and Red in another.
colorDf = peopleDf.repartition($"color",$"code") //In this case we are passing multiple columns to //repartition and this will ensure that the
//combination of these 2 column values exist in same partition.
Total 3 Partitions are created
Blue,1 - Count =2 and both these values will be in 1 Partition.
Red, 1 - Count =1 present in 1 Partition
Blue, 2 - Count =1 present in 1 Partition
When partitioning by a column, Spark will create a minimum of 200 partitions by default.
Source:
Select dataframe columns from an Array
val cols = "id,name,address"
val colsArr = cols.split(",")
df.select(colsArr.head, colsArr.tail: _*)
Overwrite specific partitions in spark dataframe write method
Directly writing to HDFS or S3 Location
df.write.mode(SaveMode.Overwrite).save("/root/path/to/data/partition_col=value")
Source: https://stackoverflow.com/questions/38487667/overwrite-specific-partitions-in-spark-dataframe-write-method
Point 1: When Spark writes parquet file to HDFS/S3 Location, Hive is unable to read the data written by Spark.
Root Cause:
This issue is caused because of different parquet conventions used in Hive and Spark. In Hive, the decimal datatype is represented as fixed bytes (INT 32). In Spark 1.4 or later the default convention is to use the Standard Parquet representation for decimal data type. As per the Standard Parquet representation based on the precision of the column datatype, the underlying representation changes.
eg: DECIMAL can be used to annotate the following types: int32: for 1 <= precision <= 9 int64: for 1 <= precision <= 18; precision < 10 will produce a warning
Hence this issue happens only with the usage of datatypes which have different representations in the different Parquet conventions. If the datatype is DECIMAL (10,3), both the conventions represent it as INT32, hence we won't face an issue. If you are not aware of the internal representation of the datatypes it is safe to use the same convention used for writing while reading. With Hive, you do not have the flexibility to choose the Parquet convention. But with Spark, you do.
Solution: The convention used by Spark to write Parquet data is configurable. This is determined by the property spark.sql.parquet.writeLegacyFormat The default value is false. If set to "true", Spark will use the same convention as Hive for writing the Parquet data. This will help to solve the issue.
use the below configuration while submitting spark job or in other words have this as a parameter in spark-submit job.
--conf "spark.sql.parquet.writeLegacyFormat=true" \
Point 2: While reading data from RDBMS in spark via val df1 = spark.sqlContext.read.format("jdbc").
partition column can be specified for parallelism. The partition column has to be integer only and not string or timestamp. Preferably it has to be Primary Key.
Point 3: If the column names in a Dataframe has space or . in between(eg: 'Account Number') then it has to be replaced with _. Below is the code:
def modifyschema(schema: StructType) =
{
val fields = schema.fields
val fieldnames = fields.map(f => f.name)
fieldnames.map(x => {
if(x.contains(" ")) x.replace(" ","_")
else if(x.contains(".")) x.replace(".","")
else x
}
)
}
val colNames = modifyschema(dfraw2.schema)
val dfraw2_renamed = dfraw2.toDF(colNames: _*)
Executors, Cores and Executor Memory
--num-executors - Executors are the JVM Executors on each Node and Multiple Evecutors can run at same time on a single Node. https://community.hortonworks.com/questions/56240/spark-num-executors-setting.html
--executor-cores - Number of cores = Concurrent tasks an executor can run. Optimum value is 5.
--executor-memory - Memory for each Executor = Allocation of Ram for each executor
Creating UDF's in Spark
UDFs transform values from a single row within a table to produce a single corresponding output value per row. For example, most SQL environments provide an UPPER function returning an uppercase version of the string provided as input.
val df = Seq(("ID1",1),("ID2",2),("ID3",3)).toDF("Name","value")
val udfFunc = (value: Int) => {value *value}
spark.udf.register("squareUDF",udfFunc)
df.select(col("Name"),callUDF("squareUDF",(col("value")))).show
Note: This UDF is available only in this session.
Source: https://blog.cloudera.com/working-with-udfs-in-apache-spark/
Parallelism in Spark while reading from RDBMS.
As like in sqoop, specify the column name, lowerbound, upperbound and numPartitions values.
if columnname = id,
lowerbound = 1
Upperbound = 1000000
numPartitions = 100
Then, the complete read operation would have parallel connections by splitting the specified columnname, lowerbound and upperbound as 100 parallel connections.
Need to have 100 tasks specified in spark.
Eg:
spark.executor.instances=10
spark.executor.cores=10
However, need to keep resource allocation in mind where specifying 10 executors with 10 cores each will eat up Lion's share of the cluster.
So make it
spark.executor.instances=5
spark.executor.cores=5
In this case 25 parallel reads will happen which is not bad and a resonable utilization of cluster resources.
Similarly, while writing number of write executors can be specified using repartition().
Source:
https://docs.databricks.com/spark/latest/data-sources/sql-databases.html#manage-parallelism
Using Log4j
1. Create a log4j.properties file with below contents,
# Root logger option
log4j.rootLogger=INFO, file, stdout
# Direct log messages to a log file
log4j.appender.file=org.apache.log4j.RollingFileAppender
log4j.appender.file.File=/u/<LINUXPATH>/logging.log
log4j.appender.file.MaxFileSize=10MB
log4j.appender.file.MaxBackupIndex=10
log4j.appender.file.layout=org.apache.log4j.PatternLayout
log4j.appender.file.layout.ConversionPattern=%d{yyyy-MM-dd HH:mm:ss} %-5p %c{1}:%L - %m%n
# Direct log messages to stdout
log4j.appender.stdout=org.apache.log4j.ConsoleAppender
log4j.appender.stdout.Target=System.out
log4j.appender.stdout.layout=org.apache.log4j.PatternLayout
log4j.appender.stdout.layout.ConversionPattern=%d{yyyy-MM-dd HH:mm:ss} %-5p %c{1}:%L - %m%n
log4j.appender.file.Append=false
log4j.appender.file.ImmediateFlush=true
log4j.appender.file.Threshold=INFO
2. @transient lazy val logger = Logger.getLogger("AppName")
PropertyConfigurator.configure("log4j.properties")
now, we can use
logger.warn("Warning msg")
logger.error("Error msg")
logger.info("Info msg")
3. In spark submit command
spark-submit --class xyz --files /<PATH>/log4j.properties "-Dlog4j.configuration=file:/<PATH>/log4j.properties"
Spark Submit command to launch job in cluster mode
spark-submit --class namespace.XYZClass --files /<PATH>/log4j.properties#log4j.properties,/<CONFFilePath>/unit_AppConffile.conf#unit_AppConffile.conf /<PATH>/App.jar unit_AppConffile.conf
How Spark Shuffle works:
Example of word count:
rdd.flatMap(_.split(' ')).map((_, 1)).reduceByKey((x, y) => x + y).filter(_._1 > 100)
Well, after the initial map stages complete, depending on your shuflle manager, each row is either hashed by the key or sorted and put into a file on disk, on the machine that it was sourced from. Then that executors lets something called the ShuffleManager know that it currently has a block of data corresponding to the given key. The ShuffleManager keeps track of all keys/locations and once all of the map side work is done. The next stage starts, and the executors each reach out to the shuffle manager to figure out where the blocks for each of their keys live. Once they know where those blocks live, each executor will reach out to the corresponding executor to fetch the data and pull it down to be processed locally. To enable this, all the executors run a Netty server which can serve blocks that are requested from that specific executor.
So to recap, it proceeds as follows:
Map operations pipelined and executed
Map side shuffle operations performed (map side reduce, etc)
Map side blocks written to disk and tracked within the ShuffleManager/BlockManager
Reduce stage begins to fetch keys and blocks from the ShuffleManager/BlockManager
Reduce side aggregate takes place.
Next set of map/shuffle stages takes place, repeating from step #1.
Source:
http://hydronitrogen.com/apache-spark-shuffles-explained-in-depth.html
Map() vs flatMap()
Both map() and flatmap() takes single element to process.
When map() is applied on an input RDD, then the output of this function will contain same number of rows as input RDD.
Output of flatMap() can have 0,1 or multiple elements as output and it need not be same as input RDD. So, this is called flattened map.
Spark Architecture or Spark Execution Model:
https://developer.hpe.com/blog/how-spark-runs-your-applications/
Partition size in spark
By default HDFS Block size is the Spark partition size
val srcDf = spark.read.option("header",true).csv("file1")
val sinkDf = spark.read.option("header",true).csv("file2")
val df_asSrc = srcDf.as("dfSrc")
val df_asSink = sinkDf.as("dfSink")
val joined_df = df_asSrc.join(df_asSink , col("df_Src.id") === col("dfSink.id") && col("df_Src.subject") === col("dfSink.subject"), "inner")
joined_df .show
joined_df.select(col("df_Src.id"), col("df_Src.subject"), col("df_Sink.subject")).show
joined_df .select(col("df_Src.*")).show
Partitioning Pruning:
Available from Spark 3 onwards and by default
spark.sql.optimizer.dynamicPartitionPruning.enabled=true
Partition pruning is a query optimization technique in Apache Spark that reduces the amount of data read during query execution. It does this by only scanning partitions that are relevant to the query.
How it works
Partitioning: Data is divided into subsets based on specific columns.
Filter/predicate push down: Filters from the WHERE clause are applied early in the query execution plan.
Partition pruning: Only the partitions that contain data that satisfy the predicate are scanned.
Types of partition pruning
Static partition pruning: Partition filters are determined at query analysis time.
Dynamic partition pruning (DPP): Partition filters are determined at runtime, based on the actual values returned by some of the predicates.
What is the difference between Spark's Partition Pruning and Predicate Pushdown?
The difference is about who applies the optimization, where the optimization is applied and which data sources it can be applied to.
Partition pruning is applied by Spark itself before it delegates to a data source handling the file format. It's only applicable in the case of file-based formats as data sources don't have the concept of partition discovery (yet).
Predicate push down delegates filtering of rows to the data source responsible for handling a particular format (Spark's term for a type of data source). Predicate push down is available for both file-based and non-file-based sources, e.g., RDBMS and NoSQL databases.
UDF to convert column values in String to TimeStamp
def convertTimeStamp(ts: String): Timestamp ={
val timestamp = new Timestamp(new SimpleDateFormat("yyyyMMddHH:mm:ss").parse(ts).getTime());
timestamp
}
def applyTimeStampFormat(srcDf: DataFrame, columnName: String): DataFrame = {
val convertTimeStamp = udf[Timestamp, String](this.convertTimeStamp)
val srcDF = srcDf.withColumn(s"${columnName}_Tmp", convertTimeStamp(col(columnName)))
.drop(columnName).withColumnRenamed(s"${columnName}_Tmp", columnName)
srcDF
}
calling function:
val df = applyTimeStampFormat(srcDf, "TS_COLNAME")
Concatenating 2 Columns:
If we want to concat data between multiple columns, separated by a pipe and create new column, then use below code,
srcdf has columns id and name
import org,apache,spark.sql.types._
val selection = srcdf.columns.map(col)
val finaldf = srcdf.withColumn("row_key", concat_ws("|", selection: _*).cast(StringType))
This will create a new column row_key with values as 1|Leela.
RepartitionByRange
Consider a case where the data is Skewed which means if we have a record set of 10,000 records and is stored in 3 partitions and partition 1 has 7,000 records and the other 2 partitions has the remaining 3000 records. Or typically the data is not evenly distributed among the partitions.
To, distribute data evenly among all the partitions, use repartitionByRange API. This function takes 2 input parameters numberOf Partitions and ColumnName(s) on which the data has to be divided.
Eg: repartitionByRange ( 5, col("AccountId"))
Same can be achieved via bucketBy(), but repartitionByRange gives better results
Difference between RepartitionByRange vs Repartition
RepartitionByRange would repartition such that the data in each of the partition would be in a sequence. Data in each partition would be as below:
("10,11,12,13,14", "15,16,17,18", "19,20,21,22")In case of repartition it could be,
("10,14,17,18,21,22", "13,19,20", "11,12,15,16")
Difference between Spark repartition() vs partitionBy()
PySpark repartition() is a DataFrame method that is used to increase or reduce the partitions in memory and when written to disk, it create all part files in a single directory.
PySpark partitionBy() is a method of DataFrameWriter class which is used to write the DataFrame to disk in partitions, one sub-directory for each unique value in partition columns.
Bucketing:
Before we discuss on Bucketing, lets first see how a Join happens in Spark. Below are the 3 Stages happen for the records for table 1 when it joins with table. Same activity happens in table 2 as well.
1. Shuffle
2. Sort
3. Merge
Out the above Shuffle is costly operation as the records need to move between the nodes.
Bucketing helps in minimizing Shuffle.
Bucketing column needs to be the column which is not suitable for Partitioning where it can result in numerous partitions and lot of small files and are used in Join. Eg customerId
numBuckets = 3
salesData.write.bucketBy(numBuckets, "customerId").sortBy("transactionId").saveAsTable("Orders")
Above statement creates 3 buckets and would distribute records on hash(customerId)%numBuckets.
Note: Bucketed data can only be saved as a table and not files in HDFS or S3. So, only .saveAsTable or in ab existing Hive table. Reason, the table maintains information that these are the 3 files generated as part of Bucketing operation on this column. While reading the data, the bucketed info is fetched from the table.
Consider that in table 2 the column to be joined is also bucketed. Here is the key, when the Join operation happens, the 2 bucketed files those hold same keys are processed by the same executor. So here, if there are 3 executors, each executor will have the bucketed file of both the tables which has same joining Ids. This would eliminate the shuffle operation and the join happens within the executor.
Another case: If the 2nd table is not bucketed, in this case the Executor will have the Bucketed file as in it's node and matching keys of the 2nd table would be shuffled. Atleast, in this case full shuffle could be avoided.
withColumn, when, otherwise usage (column comparison)
We can use the function when to validate a condition between 2 column followed by otherwise. Please see the below code:
Basic Code:
Input Set: